Publications
Chao KH, Mao A, Salzberg SL, Pertea M. Splam: a deep-learning-based splice site predictor that improves spliced alignments, Genome Biology 25, 2024
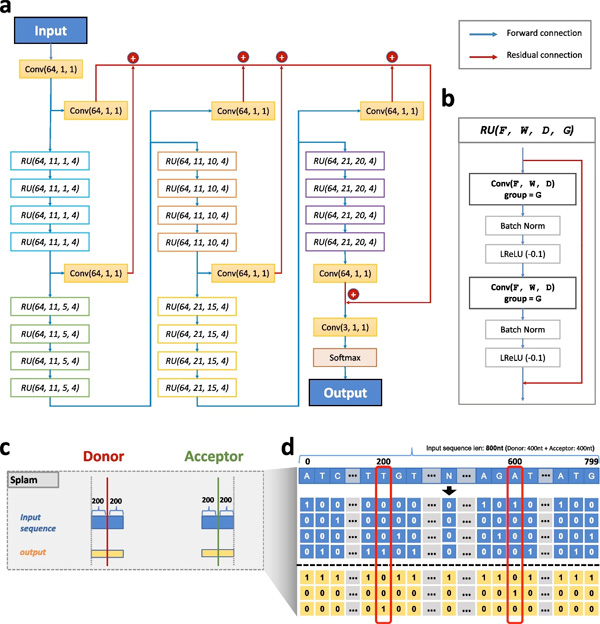
SPLAM: a deep-learning-based splice site predictor that improves spliced alignments. 2024
Pardo-Palacios FJ, et al. Systematic assessment of long-read RNA-seq methods for transcript identification and quantification, Nature Methods, 2024
Erdogdu B, Varabyou A, Hicks SC, Salzberg SL, Pertea M Detecting differential transcript usage in complex diseases with SPIT, Cell Reports Methods, 2024
Shinder I, Hu R, Ji HJ, Chao KH, Pertea M EASTR: Correcting systematic alignment errors in multi-exon genes, Nature Communications, 2023
Amaral P, et al. The status of the human gene catalogue Nature, 2023
Gihawi A, et al. Major data analysis errors invalidate cancer microbiome findings, Mbio, 2023
Varabyou A, Erdogdu B, Salzberg SL, Pertea M Investigating open reading frames in known and novel transcripts using ORFanage, Nature Computational Science, 2023
Chao KH, Zimin AV, Pertea M, Salzberg SL The first gapless, reference-quality, fully annotated genome from a Southern Han Chinese individual, G3: Genes, Genomes, Genetics, 2023
Varabyou A, Sommer MJ, Erdogdu B, Shinder I, Minkin I, Chao KH, Park S, Heinz J, Pockrandt C, Shumate A, Rincon N, Puiu D, Steinegger M, Salzberg SL, Pertea M CHESS 3: an improved, comprehensive catalog of human genes and transcripts based on large-scale expression data, phylogenetic analysis, and protein structure, Genome Biology, 2023
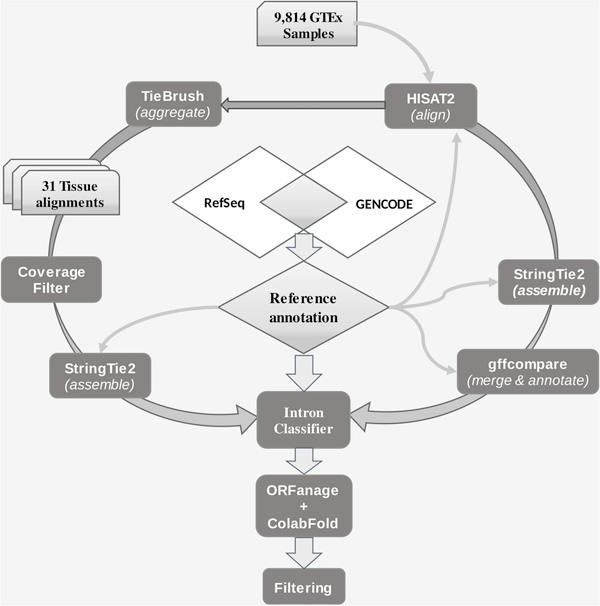
CHESS 3: an improved, comprehensive catalog of human genes and transcripts based on large-scale expression data, phylogenetic analysis, and protein structure. 2023
Sommer MJ, Cha S, Varabyou A, Rincon N, Park S, Minkin I, Pertea M, Steinegger M, Salzberg SL Structure-guided isoform identification for the human transcriptome, eLife, 2022
Tiek DM, Erdogdu E, Razaghi R, Jin L, Sadowski N, Alamillo-Ferrer C, Hogg JR, Haddad BR, Drewry DH, Wells CI, Pickett JE, Song X, Goenka A, Hu B, Goldlust SA, Zuercher WJ, Pertea M, Timp W, Cheng S-Y, Riggins RB Temozolomide-induced guanine mutations create exploitable vulnerabilities of guanine-rich DNA and RNA regions in drug-resistant gliomas, Science Advances, 2022
Shumate A, Wong B, Pertea G, Pertea M Improved transcriptome assembly using a hybrid of long and short reads with StringTie, PLOS Computational Biology, 2022
Varabyou A, Pockrandt C, Salzberg SL, Pertea M Rapid detection of inter-clade recombination in SARS-CoV-2 with Bolotie, Genetics, 2021
Varabyou A, Pertea G, Pockrandt C, Pertea M TieBrush: an efficient method for aggregating and summarizing mapped reads across large datasets, Bioinformatics, 2021
Varabyou A, Salzberg SL, Pertea M Effects of transcriptional noise on estimates of gene and transcript expression in RNA sequencing experiments, Genome research 31 (2), 301-308
Pertea G and Pertea M. GFF Utilities: GffRead and GffCompare, F1000Research 2020, 9:304 DOI:10.12688/f1000research.23297.1.
Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2 , Genome Biology, 20 (1), 1-13, 2019
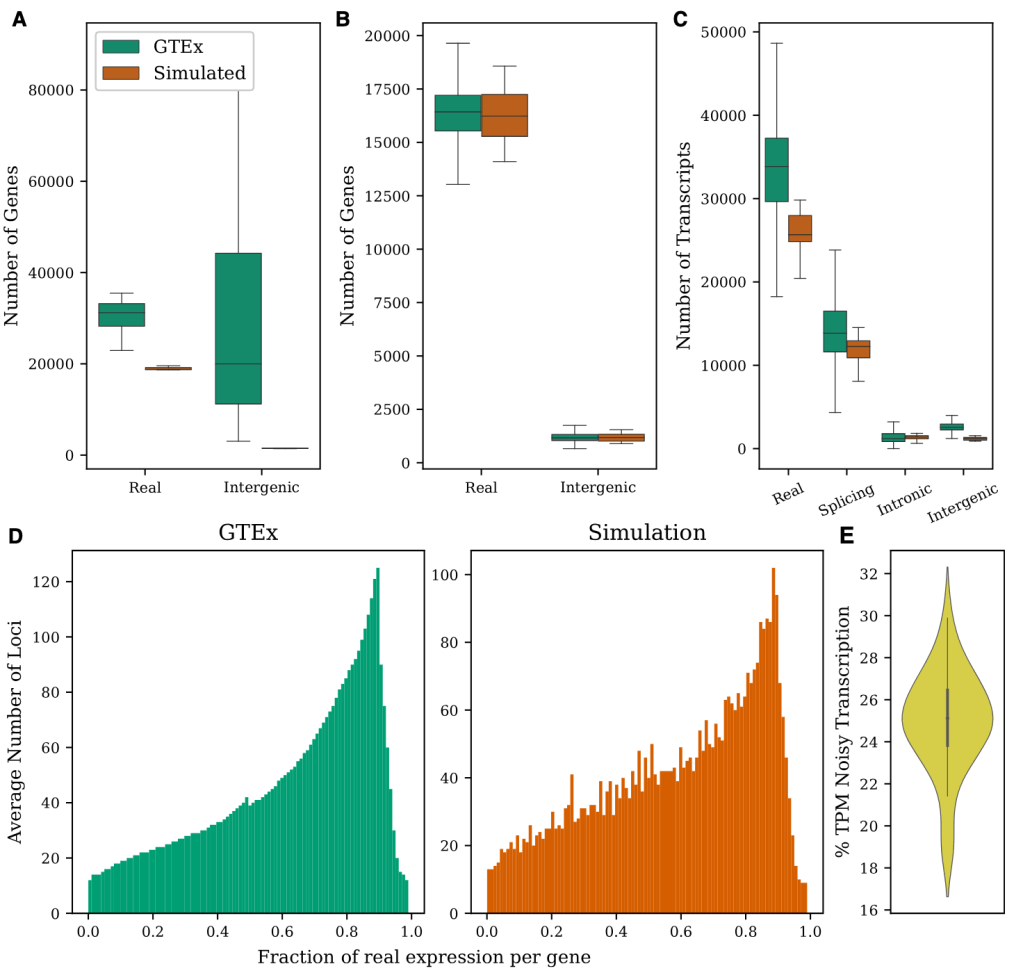
Effects of transcriptional noise on estimates of gene and transcript expression in RNA sequencing experiments, 2021.
Pertea M, Shumate A, Pertea G, Varabyou A, Breitwieser FP, Chang YC, Madugundu AK, Pandey A and Salzberg SL. CHESS: a new human gene catalog curated from thousands of large-scale RNA sequencing experiments reveals extensive transcriptional noise, Genome Biology, 2018, 19:208, doi.org/10.1186/s13059-018-1590-2
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown, Nature Protocols 11, 1650-1667 (2016), doi:10.1038/nprot.2016.095
Chang TC, Pertea M, Lee S, Salzberg SL, Mendell J. Genome-wide annotation of microRNA primary transcript structures reveals novel regulatory mechanisms, Genome Research 2015, Sep;25(9):1401-9. doi: 10.1101/gr.193607.115.
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT & Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads, Nature Biotechnology 2015, doi:10.1038/nbt.3122
Deng K, Pertea M, Rongvaux A, Durand CM, Ghiaur G, Lai J, McHugh HL, Hao H, Kumar P, Deeks SG, Siliciano JD, Salzberg SL, Flavell RA, Shan L & Siliciano RF. Broad-spectrum CTL response is required to clear latent HIV-1 due to dominance of escape mutations, NATURE 2015, 517 (7534), 381-385.
Salzberg SL, Pertea M, Fahrner JA, Sobreira N. DIAMUND: Direct Comparison of Genomes to Detect Mutations, Human Mutation 2014, 35:283288
Pertea M The Human Transcriptome: An Unfinished Story, Genes 2012, 3(3), 344-360
Pertea M, Pertea GM, Salzberg SL. Detection of lineage-specific evolutionary changes among primate species, BMC Bioinformatics 2011, 12: 274
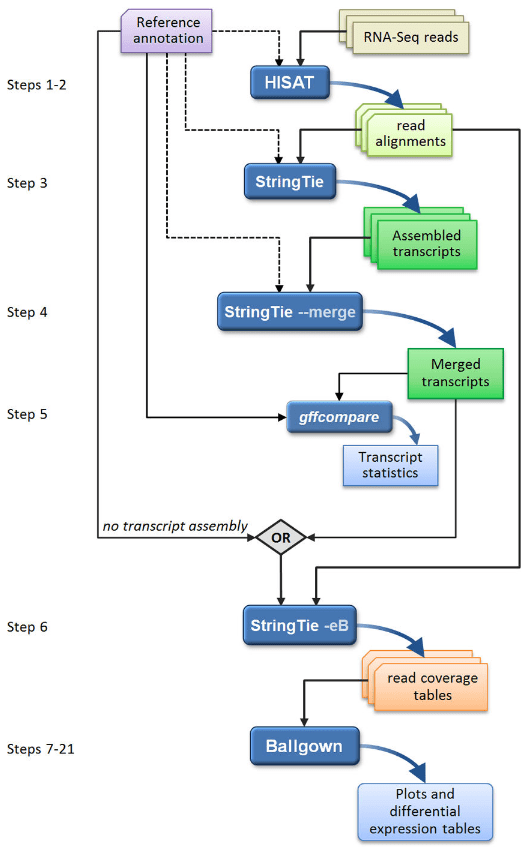
Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. 2016
Salzberg SL & Pertea M. Do-it-yourself genetic testing. Genome Biology 2010, 11:404
Pertea M, Salzberg SL. Between a chicken and a grape: estimating the number of human genes. Genome Biol. 2010 May 5;11(5):206
Pertea, M, Ayanbule, K; Smedinghoff, M, Salzberg SL. OperonDB: a comprehensive database of predicted operons in microbial genomes. NUCLEIC ACIDS RESEARCH 2009, 37: D479-D482
Haas, BJ, Salzberg, SL, Zhu, W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. GENOME BIOLOGY 2008, 9(1):R7
Pertea, M, Salzberg, SL. “Using protein domains to improve the accuracy of ab initio gene finding.” LECTURE NOTES IN COMPUTER SCIENCE, 7th International Workshop, WABI 2007 Proceedings, Vol 4645: 208-215
Pertea, M; Mount, SM; Salzberg, SL A computational survey of candidate exonic splicing enhancer motifs in the model plant Arabidopsis thaliana. BMC BIOINFORMATICS 8(159)
Allen, JE, Majoros, WH, Pertea, M, Salzberg SL. JIGSAW, GeneZilla, and GlimmerHMM: puzzling out the features of human genes in the ENCODE regions. GENOME BIOLOGY 7(S9): 1-13
Majoros WH, Pertea M, Salzberg SL. Efficient implementation of a generalized pair hidden Markov model for comparative gene finding, BIOINFORMATICS 2005 May 1;21(9):1782-8
Majoros WH, Pertea M, Delcher AL, Salzberg SL. Efficient decoding algorithms for generalized hidden Markov model gene finders, BMC BIOINFORMATICS, 2005 Jan 24;6(1):16
Majoros WH, Pertea M, and Salzberg SL, TigrScan and GlimmerHMM: two open-source ab initio eukaryotic gene-finders, BIOINFORMATICS 2004, 20(16):2878-9
Allen JE, Pertea M, Salzberg SL, Computational gene prediction using multiple sources of evidence, GENOME RES 14 (1): 142-148
StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. 2015
Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii, NATURE 419 (6906): 512-519
Pertea M, Salzberg SL, “A method to improve the performance of translation start site detection and its application for gene finding”, LECTURE NOTES IN COMPUTER SCIENCE, Second International Workshop, WABI 2002 Proceedings, Vol 2452: 210-219
Pertea M. and Salzberg S.L. Computational gene finding in plants. PLANT MOLECULAR BIOLOGY. 2002 48(1-2):39-48
Yuan Q, Quackenbush J, Sultana R, Pertea M, Salzberg SL, Buell CR. Rice bioinformatics. analysis of rice sequence data and leveraging the data to other plant species, PLANT PHYSIOL. 2001 Mar;125(3):1166-74
Pertea M, Lin X, Salzberg SL. GeneSplicer: a new computational method for splice site prediction, NUCLEIC ACIDS RES. 2001 Mar 1; 29(5):1185-90
Pertea M, Salzberg SL, Gardner MJ, Finding genes in Plasmodium falciparum,NATURE, 2000 Mar 2;404(6773):34
Salzberg SL, Pertea M, Delcher AL, Gardner MJ, Tettelin H, Interpolated Markov models for eukaryotic gene finding, GENOMICS. 1999 Jul 1;59(1):24-31.
A full list of my published work can be found at Google Scholar.